Contact
| Email: | |
|---|---|
| Phone: | +420 54949 6607 |
| Research group: | Protein-DNA Interactions - Konstantinos Tripsianes |
| Workplace: |
15. May 2018
New Software for Drug Design was a Success in an International Competition
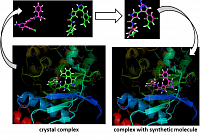
In the software competition named D3R Grand Challenge, which aims to promote technologies that will reduce the cost of drug discovery, Thomas…
8. Mar. 2018
Project for the fast and efficient method for NMR structural studies financed through an interdisciplinary fund
One of the awarded projects that will be financed through a fund supporting interdisciplinary cooperation at the Masaryk University (GAMU) belongs to…
29. Jan. 2018
New Software Developed at CEITEC MU Automates NMR Structural Studies
Press release, Brno, January 29, 2018 Researchers from CEITEC MU developed a streamlined strategy designed to meet the key objectives of NMR…
4. Apr. 2017
CEITEC MU researchers aid search for sleeping sickness cure
The prestigious journal Science has now published the results of a study examining new options for developing drugs against parasites of the Trypanosoma genus, which causes sleeping sickness and other diseases. Scientists from CEITEC at Masaryk…
Publications that are part of the Web of Science database, possibly also other publications chosen by authors.
2021
- Jurasek, M; Kumar, J; Paclikova, P; Kumari, A; Tripsianes, K; Bryja, V; Vacha, R, 2021: Phosphorylation-induced changes in the PDZ domain of Dishevelled 3. SCIENTIFIC REPORTS 11(1), doi: 10.1038/s41598-020-79398-5
- Yperman, K; Papageorgiou, AC; Merceron, R; De Munck, S; Bloch, Y; Eeckhout, D; Jiang, QH; Tack, P; Grigoryan, R; Evangelidis, T; Van Leene, J; Vincze, L; Vandenabeele, P; Vanhaecke, F; Potocky, M; De Jaeger, G; Savvides, SN; Tripsianes, K; Pleskot, R; Van Damme, D, 2021: Distinct EH domains of the endocytic TPLATE complex confer lipid and protein binding. NATURE COMMUNICATIONS 12(1), doi: 10.1038/s41467-021-23314-6
- Bousova, K; Bednarova, L; Zouharova, M; Vetyskova, V; Postulkova, K; Hofbauerova, K; Petrvalska, O; Vanek, O; Tripsianes, K; Vondrasek, J, 2021: The order of PDZ3 and TrpCage in fusion chimeras determines their properties-a biophysical characterization. PROTEIN SCIENCE 30(8), p. 1653 - 1666, doi: 10.1002/pro.4107
2019
- Pabis, M; Corsini, L; Vincendeau, M; Tripsianes, K; Gibson, TJ; Brack-Werner, R; Sattler, M, 2019: Modulation of HIV-1 gene expression by binding of a ULM motif in the Rev protein to UHM-containing splicing factors. NUCLEIC ACIDS RESEARCH , doi: doi: 10.1093/nar/gkz185
- Tripsianes, K; Schutz, U; Emmanouilidis, L; Gemmecker, G; Sattler, M, 2019: Selective isotope labeling for NMR structure determination of proteins. JOURNAL OF BIOMOLECULAR NMR , doi: doi.org/10.1007/s10858-019-00241-9
- Harnos, J; Canizal, MCA; Jurasek, M; Kumar, J; Holler, C; Schambony, A; Hanakova, K; Bernatik, O; Zdrahal, Z; Gomoryova, K; Gybel, T; Radaszkiewicz, TW; Kravec, M; Trantirek, L; Rynes, J; Dave, Z; Fernandez-Llamazares, AI; Vacha, R; Tripsianes, K; Hoffmann, C; Bryja, V, 2019: Dishevelled-3 conformation dynamics analyzed by FRET-based biosensors reveals a key role of casein kinase 1. NATURE COMMUNICATIONS 10, doi: 10.1038/s41467-019-09651-7
- Tripsianes, K; Schutz, U; Emmanouilidis, L; Gemmecker, G; Sattler, M, 2019: Selective isotope labeling for NMR structure determination of proteins in complex with unlabeled ligands. JOURNAL OF BIOMOLECULAR NMR 73(43558), p. 183 - 189, doi: 10.1007/s10858-019-00241-9
- Hanakova, K; Bernatik, O; Kravec, M; Micka, M; Kumar, J; Harnos, J; Ovesna, P; Paclikova, P; Radsetoulal, M; Potesil, D; Tripsianes, K; Cajanek, L; Zdrahal, Z; Bryja, V, 2019: Comparative phosphorylation map of Dishevelled 3 links phospho-signatures to biological outputs. CELL COMMUNICATION AND SIGNALING 17(1), doi: 10.1186/s12964-019-0470-z
2018
- Camargo, DCR; Garg, D; Buday, K; Franko, A; Camargo, AR; Schmidt, F; Cox, SJ; Suladze, S; Haslbeck, M; Mideksa, YG; Gemmecker, G; Aichler, M; Mettenleiter, G; Schulz, M; Walch, AK; de Angelis, MH; Feige, MJ; Sierra, CA; Conrad, M; Tripsianes, K; Ramamoorthy, A; Reif, B, 2018: hIAPP forms toxic oligomers in plasma. CHEMICAL COMMUNICATIONS 54(43), p. 5426 - 5429, doi: 10.1039/c8cc03097a
- Supekar, S; Papageorgiou, AC; Gemmecker, G; Peltzer, R; Johansson, MP; Tripsianes, K; Sattler, M; Kaila, VRI, 2018: Conformational Selection of Dimethylarginine Recognition by the Survival Motor Neuron Tudor Domain. ANGEWANDTE CHEMIE-INTERNATIONAL EDITION 57(2), p. 486 - 490, doi: 10.1002/anie.201708233
- Evangelidis, T; Nerli, S; Novacek, J; Brereton, AE; Karplus, PA; Dotas, RR; Venditti, V; Sgourakis, NG; Tripsianes, K, 2018: Automated NMR resonance assignments and structure determination using a minimal set of 4D spectra. NATURE COMMUNICATIONS 9, doi: 10.1038/s41467-017-02592-z
- Marcos, E; Chidyausiku, TM; McShan, AC; Evangelidis, T; Nerli, S; Carter, L; Nivón, LG; Davis, A.; Oberdorfer, G; Tripsianes, K; Sgourakis, NG; Baker, D, 2018: De novo design of a non-local β-sheet protein with high stability and accuracy. NATURE STRUCTURAL & MOLECULAR BIOLOGY , doi: 10.1038/s41594-018-0141-6
- Marcos, E; Chidyausiku, TM; McShan, AC; Evangelidis, T; Nerli, S; Carter, L; Nivon, LG; Davis, A; Oberdorfer, G; Tripsianes, K; Sgourakis, NG; Baker, D, 2018: De novo design of a non-local beta-sheet protein with high stability and accuracy. NATURE STRUCTURAL & MOLECULAR BIOLOGY 25(11), p. 1028 - +, doi: 10.1038/s41594-018-0141-6
2017
- Emmanouilidis, L; Schutz, U; Tripsianes, K; Madl, T; Radke, J; Rucktaschel, R; Wilmanns, M; Schliebs, W; Erdmann, R; Sattler, M, 2017: Allosteric modulation of peroxisomal membrane protein recognition by farnesylation of the peroxisomal import receptor PEX19. NATURE COMMUNICATIONS 8(13), doi: doi:10.1038/ncomms14635
- Dawidowski, M; Emmanouilidis, L; Kalel, V C; Tripsianes, K; Schorpp, K; Hadian, K; Kaiser, M; Mäser, P; Kolonko, M; Tanghe, S; Rodriguez, A; Schliebs, W;. Erdmann, R; Sattler, M; Popowicz, G M, 2017: Inhibitors of PEX14 disrupt protein import into glycosomes and kill Trypanosoma parasites. SCIENCE 355(6332), p. 1416 - 1420, doi: 10.1126/science.aal1807
- Camargo, DCR; Tripsianes, K; Buday, K; Franko, A; Gobl, C; Hartlmuller, C; Sarkar, R; Aichler, M; Mettenleiter, G; Schulz, M; Boddrich, A; Erck, C; Martens, H; Walch, AK; Madl, T; Wanker, EE; Conrad, M; de Angelis, MH; Reif, B, 2017: The redox environment triggers conformational changes and aggregation of hIAPP in Type II Diabetes. SCIENTIFIC REPORTS 7, doi: 10.1038/srep44041
- Supekar, S; Papageorgiou, AC; Gemmecker, G; Peltzer, R; Johansson, MP; Tripsianes, K; Sattler, M;Kaila VRK, 2017: Conformational Selection of Dimethylarginine Recognition by the Survival of Motor Neuron Tudor Domain. ANGEWANDTE CHEMIE
- PEKAŘÍK, V.; PEŠKOVÁ, M.; GURÁŇ, R.; NOVÁČEK, J.; HEGER, Z.; TRIPSIANES, K.; KUMAR, J.; ADAM, V., 2017: Visualization of stable ferritin complexes with palladium, rhodium and iridium nanoparticles detected by their catalytic activity in native polyacrylamide gels. DALTON TRANSACTIONS 46(40), p. 13690 - 5, doi: 10.1039/c7dt02818k; FULL TEXT
2016
- Valuchova, S; Fulnecek, J; Petrov, AP; Tripsianes, K; Riha, K, 2016: A rapid method for detecting protein-nucleic acid interactions by protein induced fluorescence enhancement. SCIENTIFIC REPORTS 6, doi: 10.1038/srep39653
2015
- Camargo, DCR; Tripsianes, K; Kapp, TG; Mendes, J; Schubert, J; Cordes, B; Reif, B, 2015: Cloning, expression and purification of the human Islet Amyloid Polypeptide (hIAPP) from Escherichia coli. PROTEIN EXPRESSION AND PURIFICATION 106, p. 49 - 56, doi: 10.1016/j.pep.2014.10.012
2014
- Tripsianes, K; Friberg, A; Barrandon, C; Brooks, M; van Tilbeurgh, H; Seraphin, B; Sattler, M, 2014: A Novel Protein-Protein Interaction in the RES (REtention and Splicing) Complex. JOURNAL OF BIOLOGICAL CHEMISTRY 289(41), p. 28640 - 28650, doi: 10.1074/jbc.M114.592311
- Zhang, ZY; Porter, J; Tripsianes, K; Lange, OF, 2014: Robust and highly accurate automatic NOESY assignment and structure determination with Rosetta. JOURNAL OF BIOMOLECULAR NMR 59(3), p. 135 - 145, doi: 10.1007/s10858-014-9832-4
- Tripsianes, K; Chu, NK; Friberg, A; Sattler, M; Becker, CFW, 2014: Studying Weak and Dynamic Interactions of Posttranslationally Modified Proteins using Expressed Protein Ligation. ACS CHEMICAL BIOLOGY 9(2), p. 347 - 352, doi: 10.1021/cb400723j
2013
- Kateb, F; Perrin, H; Tripsianes, K; Zou, P; Spadaccini, R; Bottomley, M; Franzmann, TM; Buchner, J; Ansieau, S; Sattler, M, 2013: Structural and Functional Analysis of the DEAF-1 and BS69 MYND Domains. PLOS ONE 8(1), doi: 10.1371/journal.pone.0054715
2012
- Mathioudakis, N; Palencia, A; Kadlec, J; Round, A; Tripsianes, K; Sattler, M; Pillai, RS; Cusack, S, 2012: The multiple Tudor domain-containing protein TDRD1 is a molecular scaffold for mouse Piwi proteins and piRNA biogenesis factors. RNA-A PUBLICATION OF THE RNA SOCIETY 18(11), p. 2056 - 2072, doi: 10.1261/rna.034181.112
- Liokatis, S; Edlich, C; Soupsana, K; Giannios, I; Panagiotidou, P; Tripsianes, K; Sattler, M; Georgatos, SD; Politou, AS, 2012: Solution Structure and Molecular Interactions of Lamin B Receptor Tudor Domain. JOURNAL OF BIOLOGICAL CHEMISTRY 287(2), p. 1032 - 1042, doi: 10.1074/jbc.M111.281303
2011
- Tripsianes, K; Madl, T; Machyna, M; Fessas, D; Englbrecht, C; Fischer, U; Neugebauer, KM; Sattler, M, 2011: Structural basis for dimethylarginine recognition by the Tudor domains of human SMN and SPF30 proteins. NATURE STRUCTURAL & MOLECULAR BIOLOGY 18(12), p. 1414 - U136, doi: 10.1038/nsmb.2185
2010
- Tripsianes, K; Sattler, M, 2010: Repeat Recognition. STRUCTURE 18(10), p. 1228 - 1229, doi: 10.1016/j.str.2010.09.006
- Meyer, NH; Tripsianes, K; Vincendeau, M; Madl, T; Kateb, F; Brack-Werner, R; Sattler, M, 2010: Structural Basis for Homodimerization of the Src-associated during Mitosis, 68-kDa Protein (Sam68) Qua1 Domain. JOURNAL OF BIOLOGICAL CHEMISTRY 285(37), p. 28893 - 28901, doi: 10.1074/jbc.M110.126185
- RecQ4 – a protein hub required for proper replication and recombination and its implications in Rothmund-Thomson Syndrome, Masarykova univerzita, 2020 - 2022
- Working from distance: engineering allosteric control on PDZ3 domain selectivity from ZO-1 protein through chimera domain fusion, Grantová agentura ČR, 2019 - 2021
- Pushing the limits in automated NMR structure determination using a single 4D NOESY spectrum and machine learning methods, Masarykova univerzita, 2018 - 2020
- From protein sequence to NMR structure in a matter of days using CHAINS/Rosetta (MUNI/E/0086/2017), Masarykova univerzita, 2017 - 2017
- Molekulární stavba protein-DNA komplexů zapojených v opravě nukleotidových sestřihů, Grantová agentura ČR, 2015 - 2017
- Molekulární stavba protein-DNA komplexů zapojených v opravě nukleotidových sestřihů (GA15-22380Y), Grantová agentura ČR, 2015 - 2017
- Structural studies of Nucleotide Excision Repair for drug development targeting protein-DNA interactions, Evropská unie, 2013 - 2017